Impact of the viral RNA epitranscriptome during infection (ViroMOD)
Funding: Fonds régional de coopération pour la recherche-APP 2021-2023, PI: Yuri Motorin (Université de Lorraine) ; Co-PI: Carine Meignin (Université de Strasbourg) and Laurent Andreoletti (66,6K euros from Feder funds).
Collaborations:
- IMoPA (Ingénierie Moléculaire et Physiopathologie Articulaire) - UMR 7365 CNRS – Université de Lorraine
- M3i Modèles insectes d’immunité innée – UPR 9022 – Université de Strasbourg
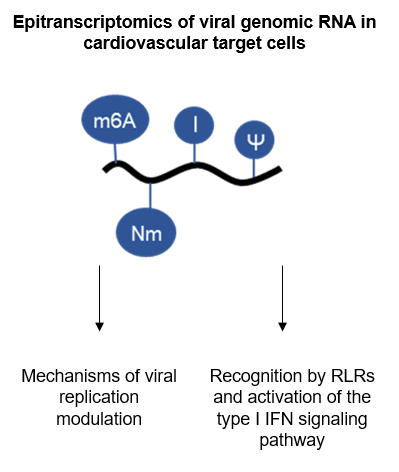
RNA modifications, commonly referred to as epitranscriptome, are observed on specific residues of cellular RNAs and play a key role for their maturation and functionality. Among more than one hundred and fifty described chemical modifications, methylation of nucleotides at different positions is the most abundant. In the last decade, the development of new detection approaches by high-throughput genome-wide sequencing has led to the identification of mRNA methylation providing a new level of regulation of gene expression.
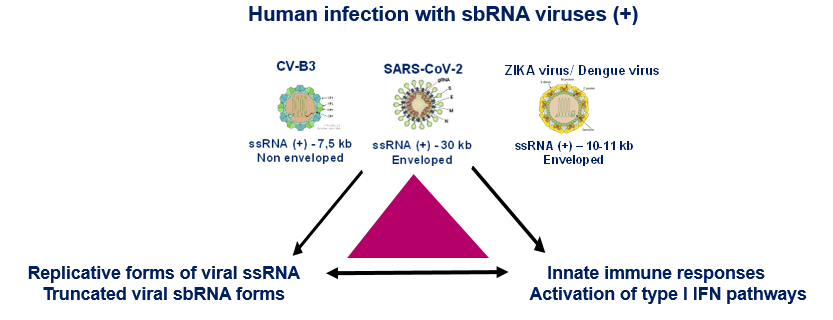
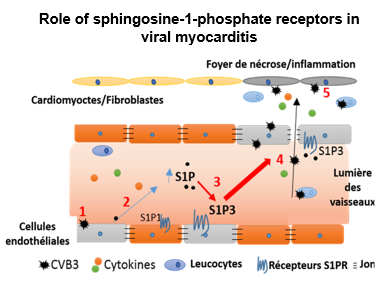
However, very few studies have been devoted so far to viral RNA modifications and, despite the identification of some RNA modifications present in viruses, little is known about their roles in virus infection and propagation. Some modifications of viral RNAs appear today fundamental for the regulation of arboviruses (ZIKA and Dengue) and other viruses. This ambitious project has an integrative dimension, since it combines the mapping of the epitranscriptome of viral RNAs with genetic and molecular virology approaches.
The ViroMOD project brings together partners in Lorraine, Alsace and Champagne-Ardenne who are scientists with international visibility and diverse backgrounds including RNA biologists (IMoPA, Nancy), experts in molecular virology and insect genetics (UPR9022, Strasbourg) and in medical virology (UMR-S 1320, ex-EA-4684, Reims). The ViroMOD project will generate complementary expertises by integrating different technological approaches and biological models.
The project has three scientific objectives: to map post-transcriptional modifications of genomic RNAs (gRNAs) of positive polarity for CVB3 and SARS-Cov-2 viruses; to study the impact of gRNA modifications on viral genome replication and maturation; and to study the detection of modified viral RNAs by the host innate immune system
In the field of molecular virology, the comprehensive characterization of the epitranscriptome of viral RNAs will be crucial for a better understanding of the defense mechanisms against these infections. Modulation of viral RNA modifications may help to restore the efficiency of recognition mechanisms as well as the elimination of the virus, or, at least, limit its spread to a very large extent.
Key words: Infection, molecular virology, epitranscriptomics, RNA modification